For the past few years, we have created innovate methods to improve earlier stages of the antibiotic discovery process. This includes a mass spectrometry-based bioinformatics tool (IDBac) to minimize redundancy of NP production between isolates, and a dual sided agar plate assay (DAPA) which allows microorganisms to compete on opposing sides of a solid support in individual wells. To integrate these advances into a single pipeline, we developed a new Environment to Bioassay antibiotic discovery approach that combines high-throughput robotics with DAPA and IDBac to rapidly select, screen, and prioritize antibiotic-producing bacteria. This framework allows us to accomplish many stages of the microbial drug discovery pipeline directly from bacterial cell mass grown on multiwell plates in a semi-automated fashion and offers an advantage in terms of scale and capacity. This project has been integrated with educational outreach efforts in partnership with community centers from underserved areas of Chicago, such as the James Jordan Boys & Girls Club. There are three central components to the program – field work, applied science experiments, and environmental literacy – with the goal of inspiring students from marginalized backgrounds to become the next cohort of university STEM majors. Overall, we aim to demonstrate 1) the feasibility and efficiency of a new antibiotic discovery approach, and 2) that graduate-level research can be successfully integrated with community partnerships.
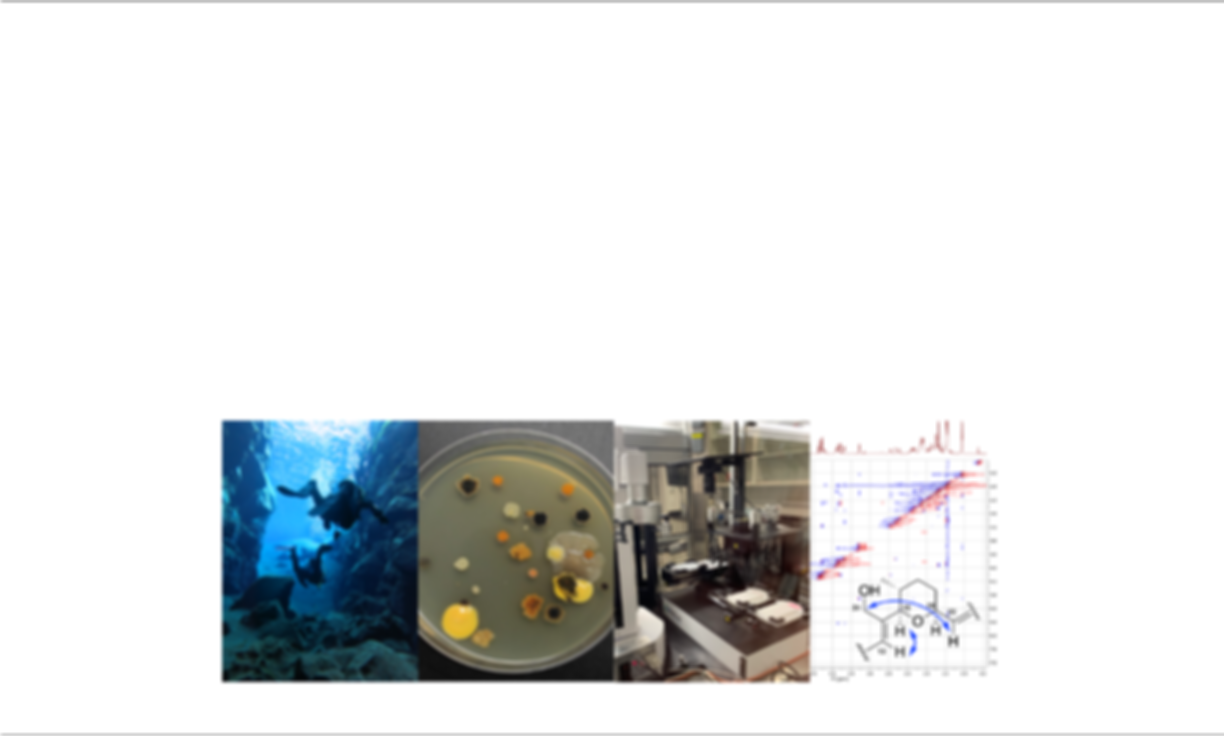
With the growing threat of antimicrobial resistance, there is an urgent need to discover new antibiotics. Microorganisms are a major source of antibacterial drugs, but a typical pipeline employed for microbial drug discovery is highly time consuming, labor intensive, and often results in re-isolation of known antibiotics. For the past few years, we have created innovate methods to improve earlier stages of the antibiotic discovery process. This includes a mass spectrometry-based bioinformatics tool (IDBac) to minimize redundancy of NP production between isolates, and a dual sided agar plate assay (DAPA) which allows microorganisms to compete on opposing sides of a solid support in individual wells. To integrate these advances into a single pipeline, we developed a new Environment to Bioassay antibiotic discovery approach that combines high-throughput robotics with DAPA and IDBac to rapidly select, screen, and prioritize antibiotic-producing bacteria. This framework allows us to accomplish many stages of the microbial drug discovery pipeline directly from bacterial cell mass grown on multiwell plates in a semi-automated fashion and offers an advantage in terms of scale and capacity. This project has been integrated with educational outreach efforts in partnership with community centers from underserved areas of Chicago, such as the James Jordan Boys & Girls Club. There are three central components to the program – field work, applied science experiments, and environmental literacy – with the goal of inspiring students from marginalized backgrounds to become the next cohort of university STEM majors. Overall, we aim to demonstrate 1) the feasibility and efficiency of a new antibiotic discovery approach, and 2) that graduate-level research can be successfully integrated with community partnerships.